Metaviz Docker Containers
Setup and Installation
Metaviz UI along with the webservices and the graph database are available as docker containers for self-hosting the application. We use docker-compose to manage these containers and the code is available at Metaviz Docker. The following provides the list of containers we use and the url to access them
| App | Docker Container Name | Port | URL to access |
|---|---|---|---|
| Graph Database | db | 7474 | http://localhost:7474 |
| Web Services | services | 5000 | http://localhost:5000 |
| Metaviz UI | ui | 5500 | http://localhost:5500 |
| BioConductor | bioc | - | - |
For installing docker and docker-compose, Please follow the instructions at
Docker: https://docs.docker.com/engine/installation/
Docker Compose: https://docs.docker.com/compose/install/
We provide an easy to use python script metaviz.py (available at Metaviz Docker) to manage the Metaviz Docker instances. The following commands are available
| Command | Description | Params |
|---|---|---|
| build | builds all the metaviz docker containers. | None |
| serve | builds and runs the docker containers | None |
| restart | restarts metaviz docker containers | None |
| add | Load a biom file to the docker instance. | <file_location> <datasource_name> |
python metaviz.py -h displays the available commands and params required to run.
Adding Data from a Biom File to the Metaviz Docker Instance
MetavizR is an R-package to manage metagenomic data from locally hosted biom files and visualize the data using the Metaviz UI. We provide functions as part of the R-package to import data from Biom Files into a Neo4J Database. The BioConductor docker container helps with this process. It installs the metavizR package and its dependencies and calls the functions to import data into the database.
The following command loads the metagenomic dataset from a biom file (test_file.biom) and import it to the database as
python metaviz.py add <test_file.biom> <dataset_name>
Note: The import process runs in the background and to check if it is done, please use the command docker-compose logs bioc. The data won’t be available until the bioc container is done with the import.
Accessing Neo4j Docker instance
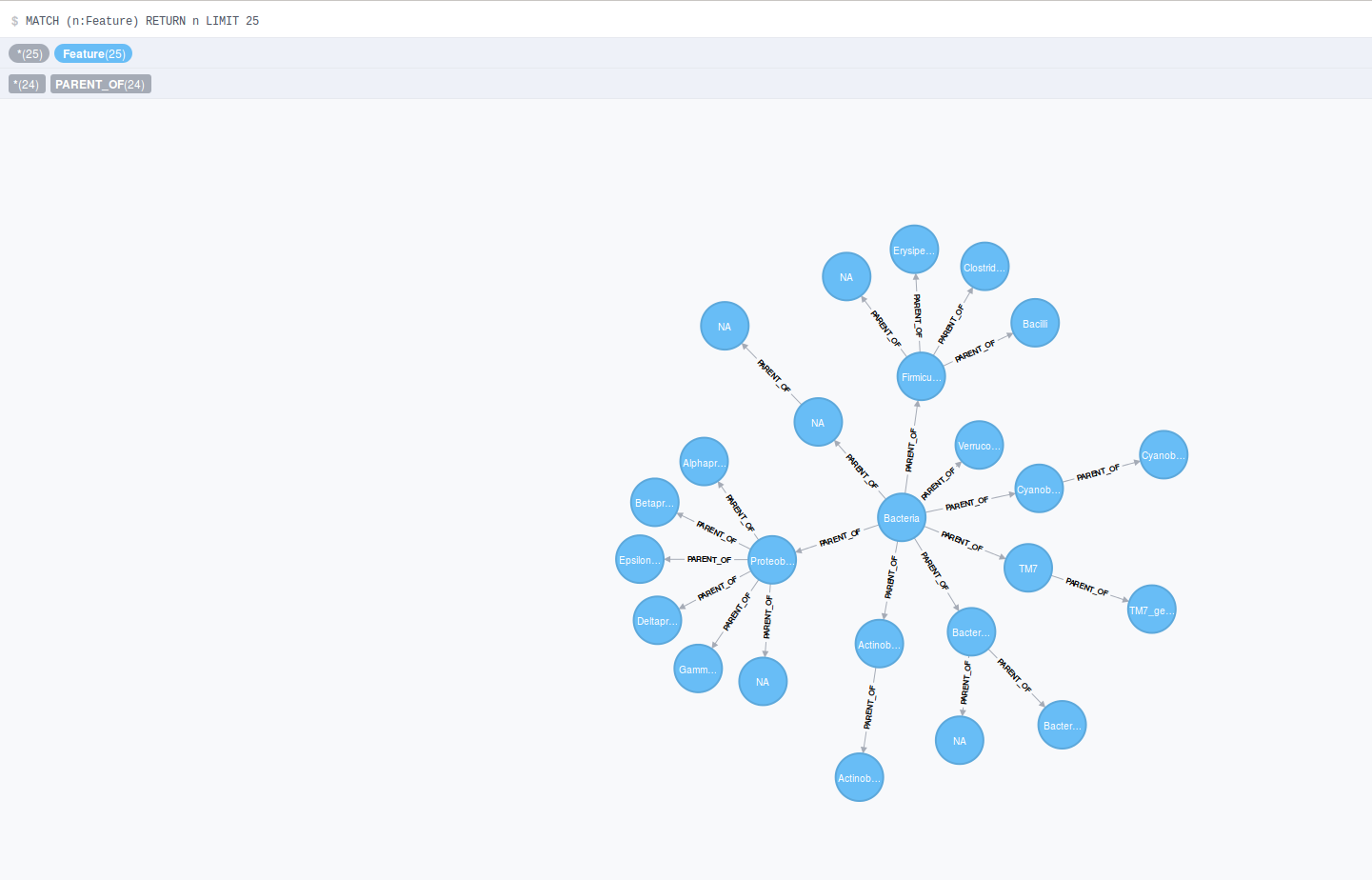
Once the data is loaded successfully, neo4j has a built-in UI to explore the graph datasets and can be accessed at http://localhost:7474 We first show the feature nodes and hiearchy connected by edges.
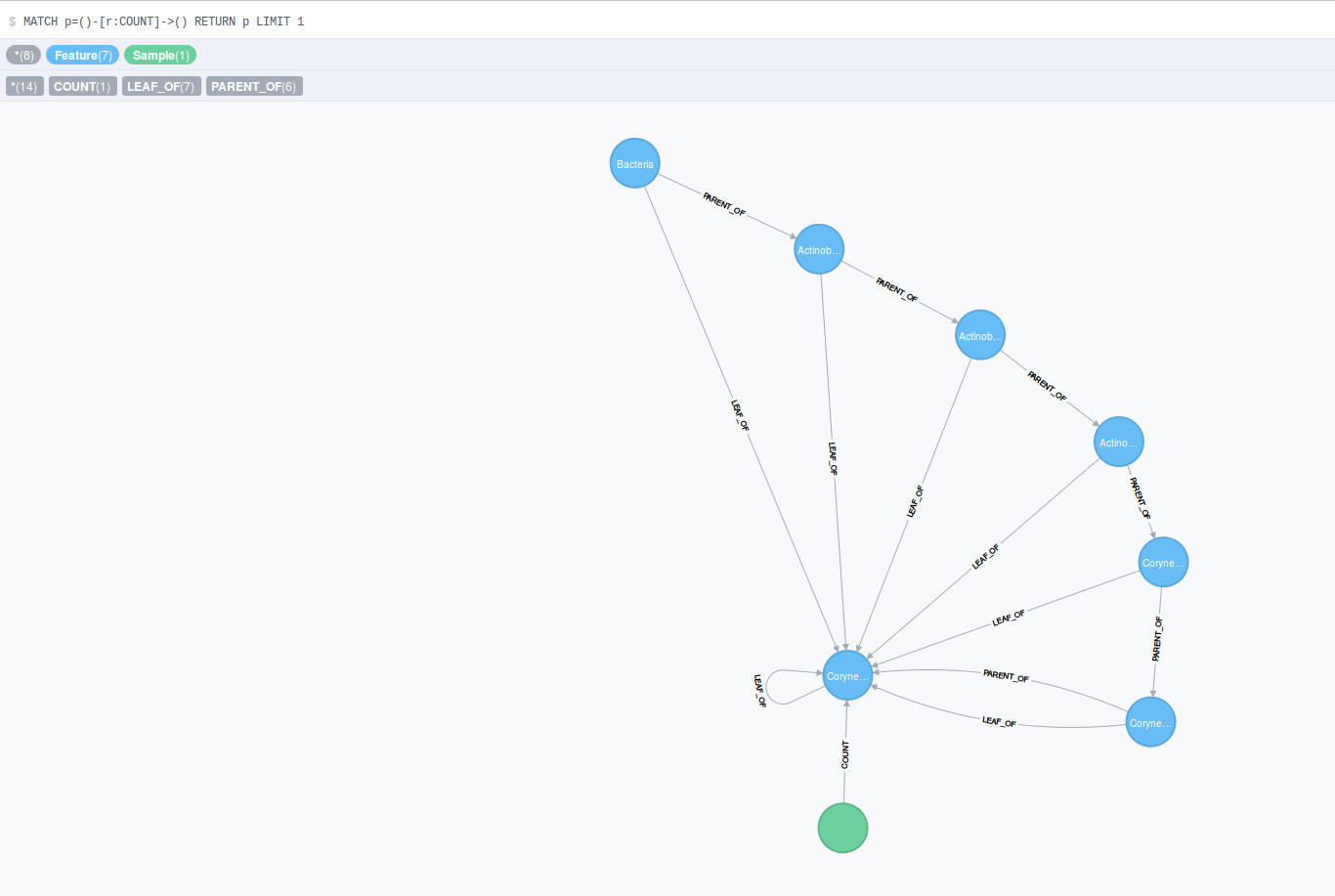
Then we show one path through the feature hierarchy to a leaf node and the count value of one sample as the edge to that leaf node. This denotes the observed counts for that feature in that sample.
Visualizing Metagenomic Data using metaviz UI
To visualize the data loaded through a biom file using metaviz, please follow the instructions at Metaviz Tutorial using msd16s Dataset.
Highlights of the features available through the Metaviz Application, please visit Metaviz Features.